The article was written by Guanao Yan, Ph.D. pupil of Statistics and Knowledge Science at UCLA. Guanao is the primary creator of the Nature Communications evaluate article [1].
Spatially resolved transcriptomics (SRT) is revolutionizing Genomics by enabling the high-throughput measurement of gene expression whereas preserving spatial context. In contrast to single-cell RNA sequencing (scRNA-seq), which captures transcriptomes with out spatial location info, SRT permits researchers to map gene expression to express areas inside a tissue, offering insights into tissue group, mobile interactions, and spatially coordinated gene exercise. The rising quantity and complexity of SRT information necessitate the event of strong statistical and computational strategies, making this discipline extremely related to information scientists, statisticians, and machine studying (ML) professionals. Methods akin to spatial statistics, graph-based fashions, and deep studying have been utilized to extract significant organic insights from these information.
A key step in SRT evaluation is the detection of spatially variable genes (SVGs)—genes whose expression varies non-randomly throughout spatial areas. Figuring out SVGs is essential for characterizing tissue structure, practical gene modules, and mobile heterogeneity. Nonetheless, regardless of the fast improvement of computational strategies for SVG detection, these strategies range broadly of their definitions and statistical frameworks, resulting in inconsistent outcomes and challenges in interpretation.
In our current evaluate printed in Nature Communications [1], we systematically examined 34 peer-reviewed SVG detection strategies and launched a classification framework that clarifies the organic significance of various SVG sorts. This text supplies an outline of our findings, specializing in the three main classes of SVGs and the statistical ideas underlying their detection.

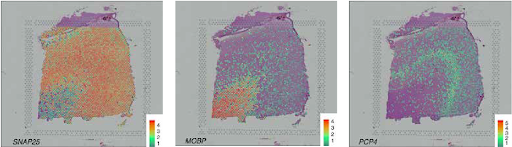
SVG detection strategies intention to uncover genes whose spatial expression displays organic patterns moderately than technical noise. Primarily based on our evaluate of 34 peer-reviewed strategies, we categorize SVGs into three teams: Total SVGs, Cell-Kind-Particular SVGs, and Spatial-Area-Marker SVGs (Determine 2).

Strategies for detecting the three SVG classes serve totally different functions (Fig. 3). First, the detection of total SVGs screens informative genes for downstream analyses, together with the identification of spatial domains and practical gene modules. Second, detecting cell-type-specific SVGs goals to disclose spatial variation inside a cell kind and assist determine distinct cell subpopulations or states inside cell sorts. Third, spatial-domain-marker SVG detection is used to search out marker genes to annotate and interpret spatial domains already detected. These markers assist perceive the molecular mechanisms underlying spatial domains and help in annotating tissue layers in different datasets.

The connection among the many three SVG classes is determined by the detection strategies, significantly the null and various hypotheses they make use of. If an total SVG detection methodology makes use of the null speculation {that a} non-SVG’s expression is unbiased of spatial location and the choice speculation that any deviation from this independence signifies an SVG, then its SVGs ought to theoretically embody each cell-type-specific SVGs and spatial-domain-marker SVGs. For instance, DESpace [2] is a technique that detects each total SVGs and spatial-domain-marker SVGs, and its detected total SVGs should be marker genes for some spatial domains. This inclusion relationship holds true besides in excessive eventualities, akin to when a gene displays reverse cell-type-specific spatial patterns that successfully cancel one another out. Nonetheless, if an total SVG detection methodology’s various speculation is outlined for a particular spatial expression sample, then its SVGs might not embody some cell-type-specific SVGs or spatial-domain-marker SVGs.
To know how SVGs are detected, we categorized the statistical approaches into three main varieties of speculation assessments:
- Dependence Check – Examines the dependence between a gene’s expression degree and the spatial location.
- Regression Mounted-Impact Check – Examines whether or not some or all the fixed-effect covariates, as an illustration, spatial location, contribute to the imply of the response variable, i.e., a gene’s expression.
- Regression Random-Impact Check (Variance Element Check) – Examines whether or not the random-effect covariates, as an illustration, spatial location, contribute to the variance of the response variable, i.e., a gene’s expression.
To additional clarify how these assessments are used for SVG detection, we denote Y as gene’s expression degree and S because the spatial areas. Dependence take a look at is essentially the most common speculation take a look at for SVG detection. For a given gene, it decides whether or not the gene’s expression degree Y is unbiased of the spatial location S, i.e., the null speculation is:

There are two varieties of regression assessments: fixed-effect assessments, the place the impact of the spatial location is assumed to be fastened, and random-effect assessments, which assume the impact of the spatial location as random. To elucidate these two varieties of assessments, we use a linear blended mannequin for a given gene for example:

the place the response variable ( Y_i ) is the gene’s expression degree at spot ( i ), ( x_i ) ( epsilon ) ( R^p ) signifies the fixed-effect covariates of spot ( i ), ( z_i ) ( epsilon ) ( R^q ) denotes the random-effect covariates of spot ( i ), and ( epsilon_i ) is the random measurement error at spot ( i ) with zero imply. Within the mannequin parameters, ( beta_0 ) is the (fastened) intercept, ( beta ) ( epsilon ) ( R^p ) signifies the fastened results, and ( gamma ) ( epsilon ) ( R^q ) denotes the random results with zero means and the covariance matrix:

On this linear blended mannequin, independence is assumed between random impact and random errors and amongst random errors.
Mounted-effect assessments look at whether or not some or all the fixed-effect covariates ( x_i ) (depending on spatial areas S) contribute to the imply of the response variable. If all fixed-effect covariates make no contribution, then:

The null speculation

implies

Random-effect assessments look at whether or not the random-effect covariates ( z_i ) (depending on spatial areas S) contribute to the variance of the response variable VarYi, specializing in the decomposition:

and testing if the contribution of the random-effect covariates is zero. The null speculation:

implies

Among the many 23 strategies that use frequentist speculation assessments, dependence assessments and random-effect regression assessments have been primarily utilized to detect total SVGs, whereas fixed-effect regression assessments have been used throughout all three SVG classes. Understanding these distinctions is essential to deciding on the correct methodology for particular analysis questions.
Enhancing SVG detection strategies requires balancing detection energy, specificity, and scalability whereas addressing key challenges in spatial transcriptomics evaluation. Future developments ought to deal with adapting strategies to totally different SRT applied sciences and tissue sorts, in addition to extending assist for multi-sample SRT information to reinforce organic insights. Moreover, strengthening statistical rigor and validation frameworks can be essential for making certain the reliability of SVG detection. Benchmarking research additionally want refinement, with clearer analysis metrics and standardized datasets to supply strong methodology comparisons.
References
[1] Yan, G., Hua, S.H. & Li, J.J. (2025). Categorization of 34 computational strategies to detect spatially variable genes from spatially resolved transcriptomics information. Nature Communication, 16, 1141. https://doi.org/10.1038/s41467-025-56080-w
[2] Cai, P., Robinson, M. D., & Tiberi, S. (2024). DESpace: spatially variable gene detection by way of differential expression testing of spatial clusters. Bioinformatics, 40(2). https://doi.org/10.1093/bioinformatics/btae027
[/latex]