language fashions has made many Pure Processing (NLP) duties seem easy. Instruments like ChatGPT typically generate strikingly good responses, main even seasoned professionals to surprise if some jobs could be handed over to algorithms sooner reasonably than later. But, as spectacular as these fashions are, they nonetheless discover duties requiring exact, domain-specific extraction.
Motivation: Why Construct a PICO Extractor?
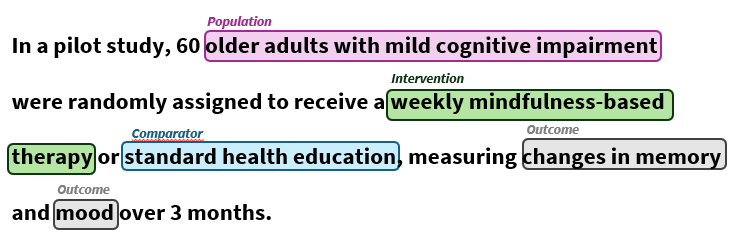
The thought arose throughout a dialog with a pupil, graduating in Worldwide Healthcare Administration, who got down to analyze future developments in Parkinson’s remedy and to calculate potential prices awaiting insurances, if the present trials flip right into a profitable product. Step one was basic and laborious: isolate PICO parts—Inhabitants, Intervention, Comparator, and End result descriptions—from operating trial descriptions revealed on clinicaltrials.gov. This PICO framework is usually utilized in evidence-based medication to construction scientific trial information. Since she was neither a coder nor an NLP specialist, she did this totally by hand, working with spreadsheets. It grew to become clear to me that, even within the LLM period, there may be actual demand for simple, dependable instruments for biomedical info extraction.
Step 1: Understanding the Information and Setting Targets
As in each information venture, the primary order of enterprise is setting clear targets and figuring out who will use the outcomes. Right here, the target was to extract PICO parts for downstream predictive analyses or meta-research. The viewers: anybody excited about systematically analyzing scientific trial information, be it researchers, clinicians, or information scientists. With this scope in thoughts, I began with exports from clinicaltrials.gov in JSON format. Preliminary discipline extraction and information cleansing supplied some structured info (Desk 1) — particularly for interventions — however different key fields have been nonetheless unmanageably verbose for downstream automated analyses. That is the place NLP shines: it permits us to distill essential particulars from unstructured textual content comparable to eligibility standards or examined medication. Named Entity Recognition (NER) permits automated detection and classification of key entities—for instance, figuring out the inhabitants group described in an eligibility part, or pinpointing final result measures inside a research abstract. Thus, the venture naturally transitioned from primary preprocessing to the implementation of domain-adapted NER fashions.

Step 2: Benchmarking Current Fashions
My subsequent step was a survey of off-the-shelf NER fashions, particularly these skilled on biomedical literature and accessible through Huggingface, the central repository for transformer fashions. Out of 19 candidates, solely BioELECTRA-PICO (110 million parameters) [1] labored straight for extracting PICO parts, whereas the others are skilled on the NER process, however not particularly on PICO recognition. Testing BioELECTRA alone “gold-standard” set of 20 manually annotated trials confirmed acceptable however removed from supreme efficiency, with explicit weak point on the “Comparator” component. This was possible as a result of comparators are not often described within the trial summaries, forcing a return to a sensible rule-based strategy, looking out straight the intervention textual content for normal comparator key phrases comparable to “placebo” or “traditional care.”
Step 3: Positive-Tuning with Area-Particular Information
To additional enhance efficiency, I moved to fine-tuning, which was made doable because of annotated PICO datasets from BIDS-Xu-Lab, together with Alzheimer’s-specific samples [2]. With the intention to stability the necessity for prime accuracy with effectivity and scalability, I chosen three fashions for experimentation. BioBERT-v1.1, with 110 million parameters [3], served as the first mannequin on account of its sturdy observe document in biomedical NLP duties. I additionally included two smaller, derived fashions to optimize for pace and reminiscence utilization: CompactBioBERT, at 65 million parameters, is a distilled model of BioBERT-v1.1; and BioMobileBERT, at simply 25 million parameters, is an extra compressed variant, which underwent a further spherical of continuous studying after compression [4]. I fine-tuned all three fashions utilizing Google Colab GPUs, which allowed for environment friendly coaching—every mannequin was prepared for testing in underneath two hours.
Step 4: Analysis and Insights
The outcomes, summarized in Desk 2, reveal clear developments. All variants carried out strongly on extracting Inhabitants, with BioMobileBERT main at F1 = 0.91. End result extraction was close to ceiling throughout all fashions. Nevertheless, extracting Interventions proved more difficult. Though recall was fairly excessive (0.83–0.87), precision lagged (0.54–0.61), with fashions incessantly tagging additional medicine mentions discovered within the free textual content—actually because trial descriptions consult with medication or “intervention-like” key phrases describing the background however not essentially specializing in the deliberate most important intervention.
On nearer inspection, this highlights the complexity of biomedical NER. Interventions sometimes appeared as quick, fragmented strings like “use of complete,” “week,” “prime,” or “tissues with”, that are of little worth for a researcher making an attempt to make sense of a compiled checklist of research. Equally, inspecting the inhabitants yielded reasonably sobering examples comparable to “% of” or “states with”, pointing to the necessity for added cleanup and pipeline optimization. On the identical time, the fashions might extract impressively detailed inhabitants descriptors, like “qualifying adults with a analysis of cognitively unimpaired, or possible Alzheimer’s illness, frontotemporal dementia, or dementia with Lewy our bodies”. Whereas such lengthy strings might be right, they are usually too verbose for sensible summarization as a result of every trial’s participant description is so particular, typically requiring some type of abstraction or standardization.
This underscores a basic problem in biomedical NLP: context issues, and domain-specific textual content typically resists purely generic extraction strategies. For Comparator parts, a rule-based strategy (matching express comparator key phrases) labored greatest, reminding us that mixing statistical studying with pragmatic heuristics is usually essentially the most viable technique in real-world purposes.
One main supply of those “mischief” extractions stems from how trials are described in broader context sections. Shifting ahead, doable enhancements embody including a post-processing filter to discard quick or ambiguous snippets, incorporating a domain-specific managed vocabulary (so solely acknowledged intervention phrases are saved), or making use of idea linking to recognized ontologies. These steps might assist make sure that the pipeline produces cleaner, extra standardized outputs.

A phrase on efficiency: For any end-user software, pace issues as a lot as accuracy. BioMobileBERT’s compact dimension translated to sooner inference, making it my most popular mannequin, particularly because it carried out optimally for Inhabitants, Comparator, and End result parts.
Step 5: Making the Device Usable—Deployment
Technical options are solely as useful as they’re accessible. I wrapped the ultimate pipeline in a Streamlit app, permitting customers to add clinicaltrials.gov datasets, swap between fashions, extract PICO parts, and obtain outcomes. Fast abstract plots present an at-a-glance view of prime interventions and outcomes (see Determine 1). I intentionally left the underperforming BioELECTRA mannequin for the person to match efficiency period so as to respect the effectivity positive aspects from utilizing a smaller structure. Though the software got here too late to spare my pupil hours of handbook information extraction, I hope it is going to profit others going through comparable duties.
To make deployment simple, I’ve containerized the app with Docker, so followers and collaborators can stand up and operating rapidly. I’ve additionally invested substantial effort into the GitHub repo [5], offering thorough documentation to encourage additional contributions or adaptation for brand new domains.
Classes Realized
This venture showcases the total journey of growing a real-world extraction pipeline — from setting clear aims and benchmarking present fashions, to fine-tuning them on specialised information and deploying a user-friendly utility. Though fashions and information have been available for fine-tuning, turning them into a very useful gizmo proved more difficult than anticipated. Coping with intricate, multi-word biomedical entities which have been typically solely partially acknowledged, highlighted the boundaries of one-size-fits-all options. The dearth of abstraction within the extracted textual content additionally grew to become an impediment for anybody aiming to determine international developments. Shifting ahead, extra targeted approaches and pipeline optimizations are wanted reasonably than counting on a easy prêt-à-porter answer.

When you’re excited about extending this work, or adapting the strategy for different biomedical duties, I invite you to discover the repository [5] and contribute. Simply fork the venture and Pleased Coding!
References
- [1] S. Alrowili and V. Shanker, “BioM-Transformers: Constructing Giant Biomedical Language Fashions with BERT, ALBERT and ELECTRA,” in Proceedings of the twentieth Workshop on Biomedical Language Processing, D. Demner-Fushman, Okay. B. Cohen, S. Ananiadou, and J. Tsujii, Eds., On-line: Affiliation for Computational Linguistics, June 2021, pp. 221–227. doi: 10.18653/v1/2021.bionlp-1.24.
- [2] BIDS-Xu-Lab/section_specific_annotation_of_PICO. (Aug. 23, 2025). Jupyter Pocket book. Scientific NLP Lab. Accessed: Sept. 13, 2025. [Online]. Obtainable: https://github.com/BIDS-Xu-Lab/section_specific_annotation_of_PICO
- [3] J. Lee et al., “BioBERT: a pre-trained biomedical language illustration mannequin for biomedical textual content mining,” Bioinformatics, vol. 36, no. 4, pp. 1234–1240, Feb. 2020, doi: 10.1093/bioinformatics/btz682.
- [4] O. Rohanian, M. Nouriborji, S. Kouchaki, and D. A. Clifton, “On the effectiveness of compact biomedical transformers,” Bioinformatics, vol. 39, no. 3, p. btad103, Mar. 2023, doi: 10.1093/bioinformatics/btad103.
- [5] ElenJ, ElenJ/biomed-extractor. (Sept. 13, 2025). Jupyter Pocket book. Accessed: Sept. 13, 2025. [Online]. Obtainable: https://github.com/ElenJ/biomed-extractor